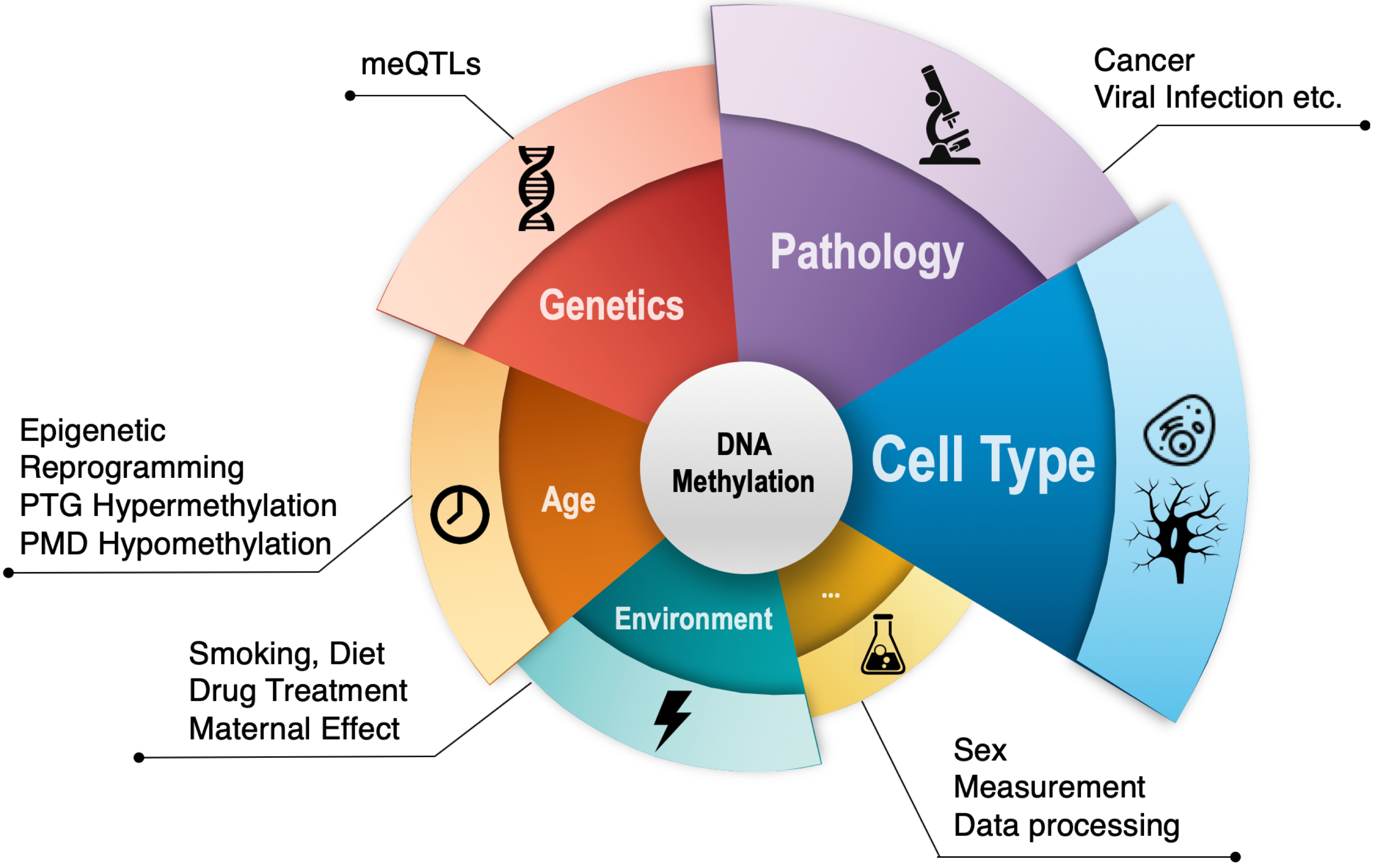
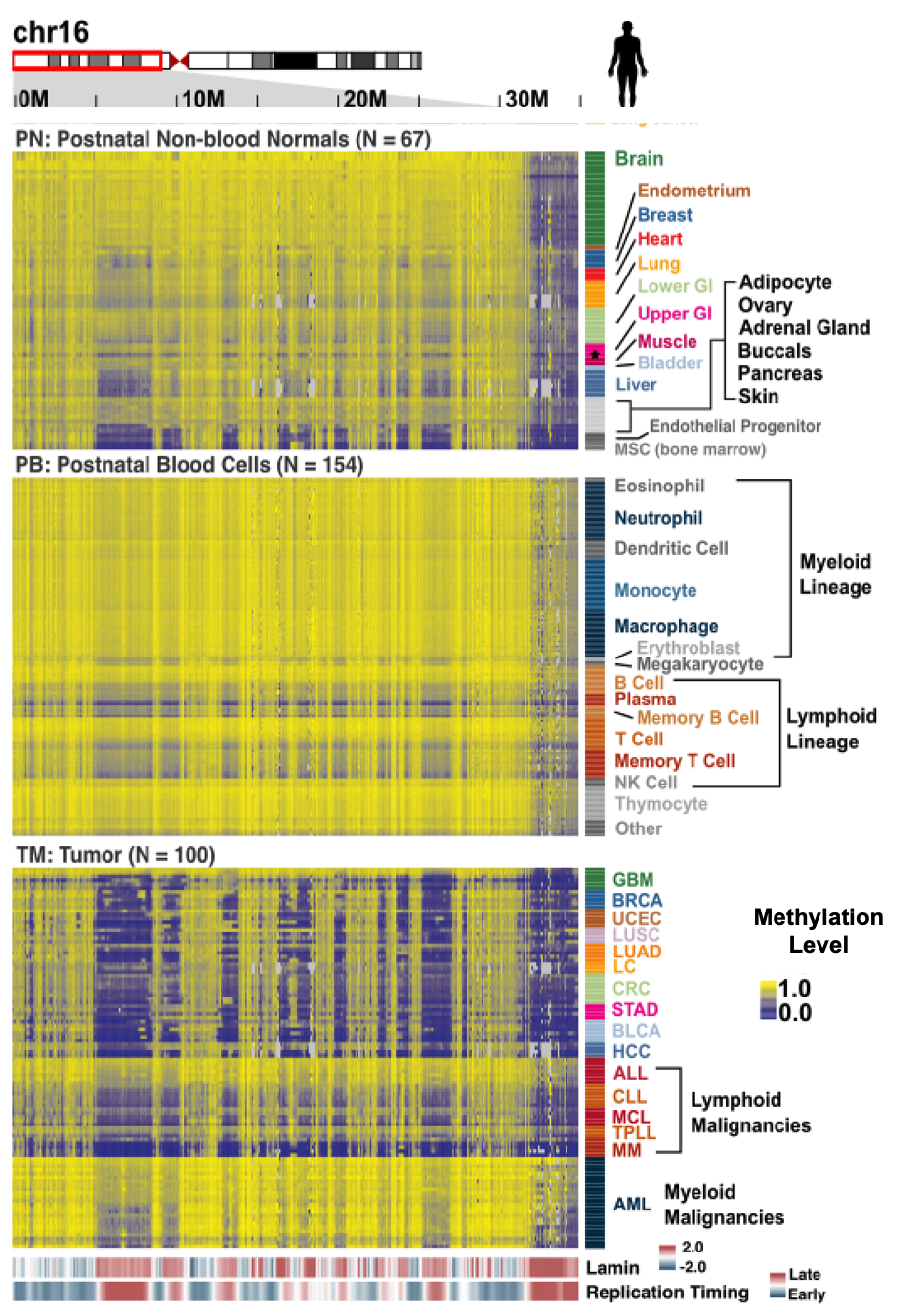
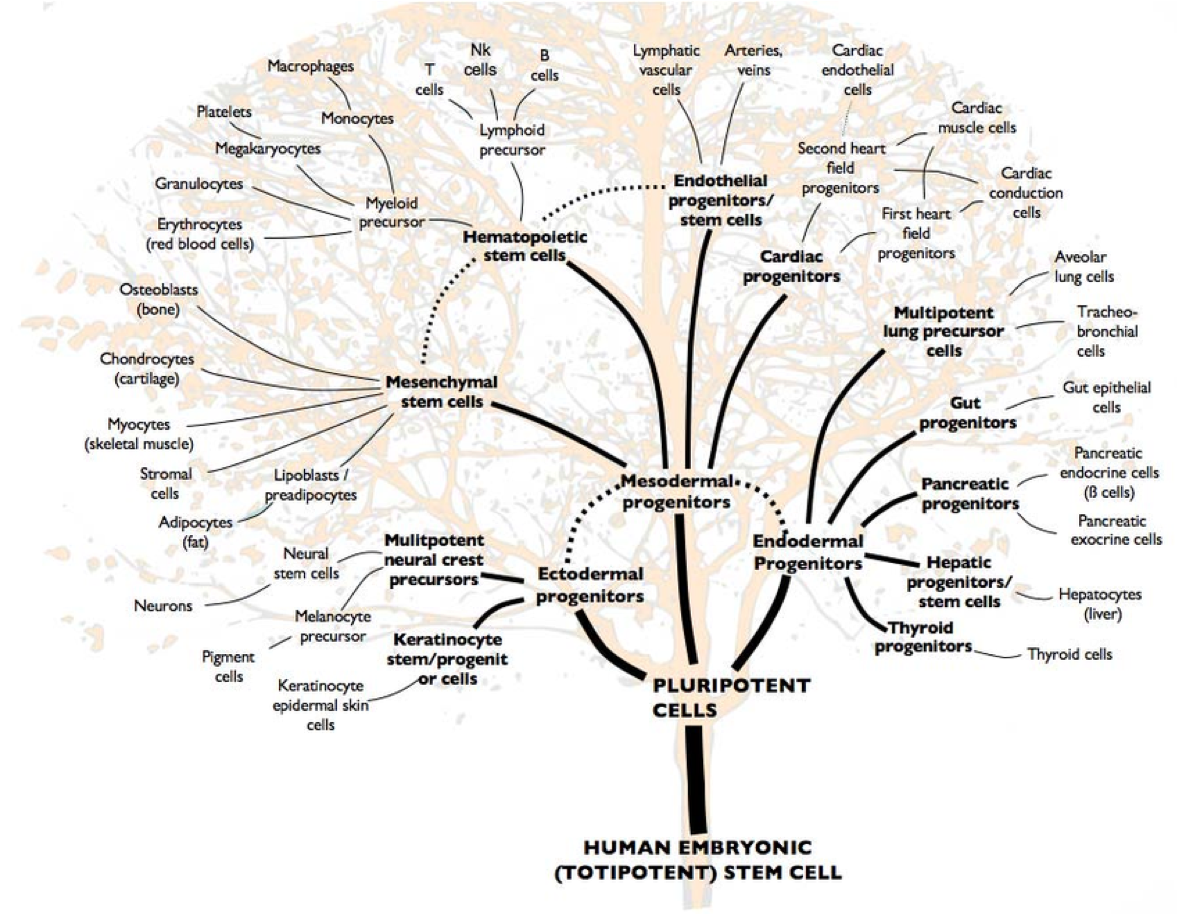
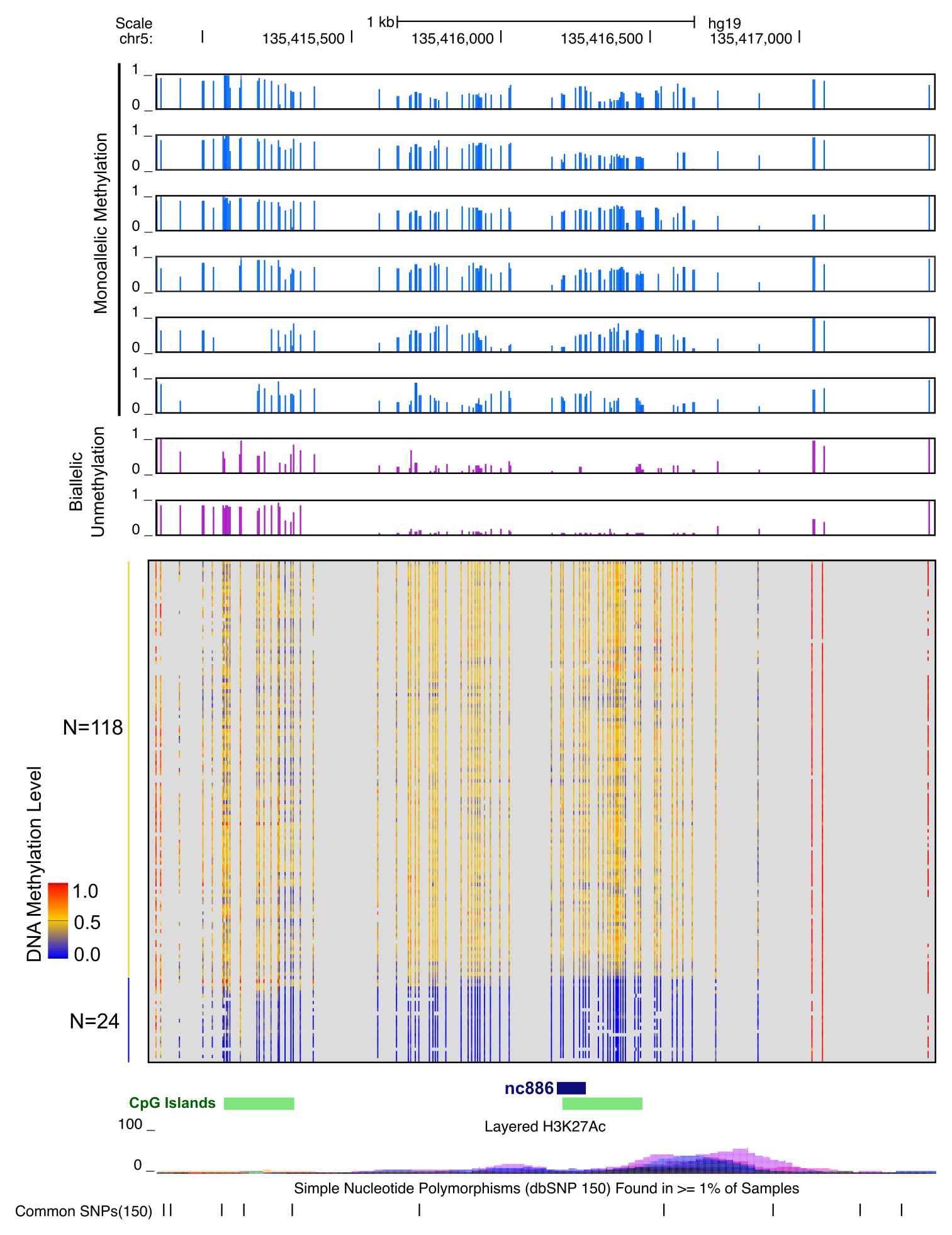
Wanding Zhou
Assistant Professor of Pathology and Lab Medicine
Postdoc in MD Anderson Cancer Center and Van Andel Institute.

Cameron Cloud
Research Technician
07/2023-
Lafayette College

Hongxiang Fu
Graduate Student
09/2023-
matric year: 09/2023

Hao Xu
Graduate Student
09/2024-
matric year: 09/2024
Alumni
- Sol Moe Lee (11/2020-11/2025): Postdoc.
- David Goldberg (12/2021-06/2025): NGG graduate student. Last position: Biomeme, Inc.
- Heqiao Zhu (09/2022-01/2025): BE graduate student.
- Zhuoran Xu (01/2024-03/2024): GCB rotation graduate student.
- Diljeet Kaur (05/2022-03/2023): on leave of absence.
- Waleed Iqbal (09/2021-11/2022): Grad student at Drexel University.
- Wubin Ding (10/2021-04/2023): Last known position: Postdoc at Salk Institute (Joe Ecker lab)
- Ethan Moyer: undergraduate research associate. Last known position: Moberg Analytics.
- Andrew Patterson (06/2021-08/2021): GCB rotation graduate student.
- Yang Wan (01/2020-10/2021): intern master student. Now at University of Pennsylvania.
- Borui Xiao (01/2020-03/2020): intern master student. Now at Hengrui Medicine Inc.