Data Science for Epigenetics
OUR MISSION
O ur central research interest is to understand tissue heterogeneity, stem cell homeostasis, and cellular aging using genomics, epigenetics, and advanced informatics. We develop informatics for DNA methylation assays, including Infinium DNA methylation microarray, bisulfite-sequencing, and single-molecule, long-read sequencing-based technologies. We are dedicated to providing sensible interpretation of various forms of epigenetic data and their integration. We create statistical inference methods to query epigenetic cell identity, cancer cell of origin, evolution, and immune microenvironment.
We focus on leveraging DNA methylation, a robust readout of the chromatin state and cell identity, to understand cellular differentiation and aging. We will integrate multi-omics data to unveil how cells translate epigenetic code into transcriptional regulation, and ultimately, phenotypical manifestation. This knowledge is essential for understanding the biological origin of organismal aging, developmental abnormalities, malignancies, and cognitive deficit. We believe data science will be a critical piece of the future of biomedical research and clinical applications. We strive to push the frontier of computational epigenetics as part of our reach to this future.
We welcome rotation students to join one or a combination of ongoing research projects listed here. All the projects can be completed virtually. The rotation student is expected to benefit from the rotation through acquiring programming skills, algorithmic thinking, conceptual grasp of the principles of statistical machine learning, and domain knowledge in epigenetics. You will be exposed to a wide range of big data analytics, exploratory visualization, scientific writing, and experience in the peer-review process. Some related prior publications can be found here. Interested students are welcome to contact me.
CHOP is an Equal Opportunity Employer. We are committed to an inclusive environment for all lab members.
OUR RESEARCH
Decoding Epigenetic Cell Identity
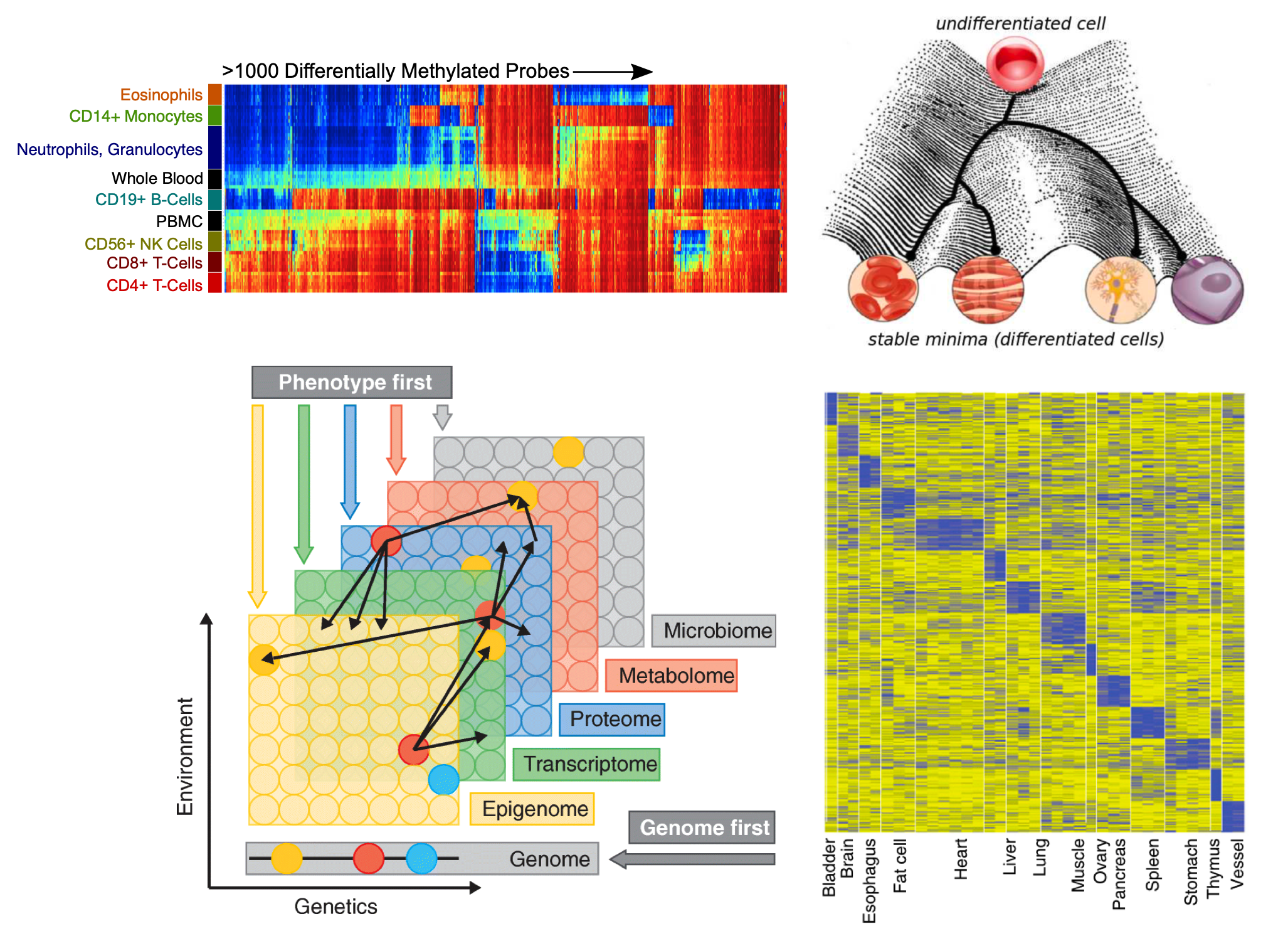
W e believe DNA methylation is one of the best markers for cell identity due to its biochemical stability and allelic presence in most cells. We measure and analyze DNA methylation signals to investigate tissue and tumor heterogeneity, tumor cell-of-origin, immune microenvironment for personalized cancer medicine, and immune therapy. We develop computational methods to perform linear deconvolution of bulk tissue DNA methylation signal. By integrating chromatin accessibility and expression profile, our research aims at reconstructing the Waddington landscape and epigenetic history of cell differential in human development and cancer.
Learn More: PNAS 2016 Immunity 2018 Science 2018
Tracing mitotic history with DNA Methylation Loss
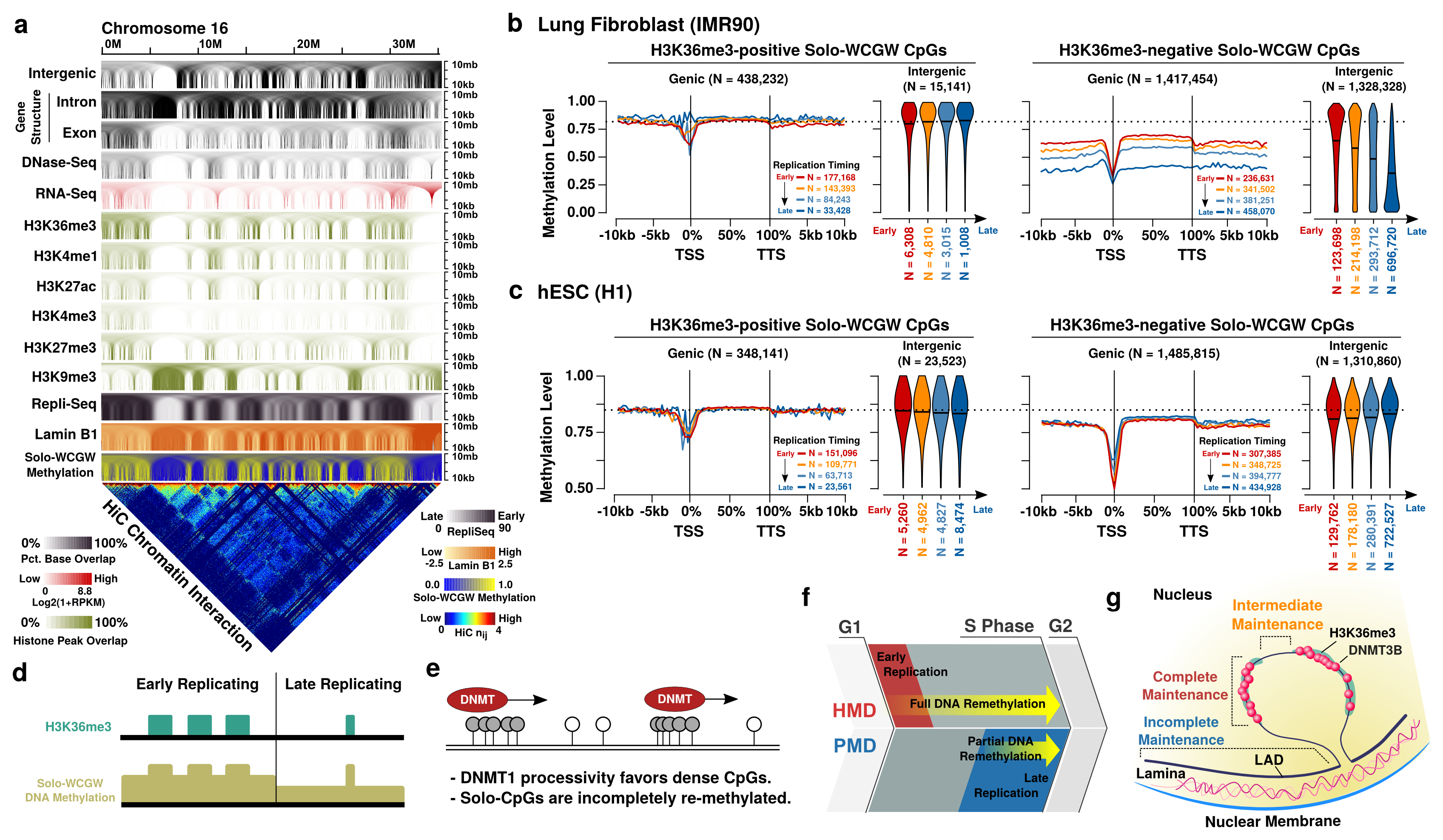
G lobal DNA methylation loss is a signature of the cancer epigenome. It occurs primarily within lamina-associated, late-replicating regions termed partially methylated domains (PMDs). Our analysis revealed previously undetected PMD hypomethylation in virtually all healthy tissues. PMD hypo-methylation increased with age, beginning during fetal development, and appeared to track the accumulation of cell divisions. In cancer, PMD hypomethylation correlated with somatic mutation density and cell cycle gene expression, suggesting its reflection of mitotic history. Late replication leads to lifelong progressive methylation loss, which acts as a biomarker for cellular aging and may contribute to oncogenesis.
Learn More: Nature Genetics 2018
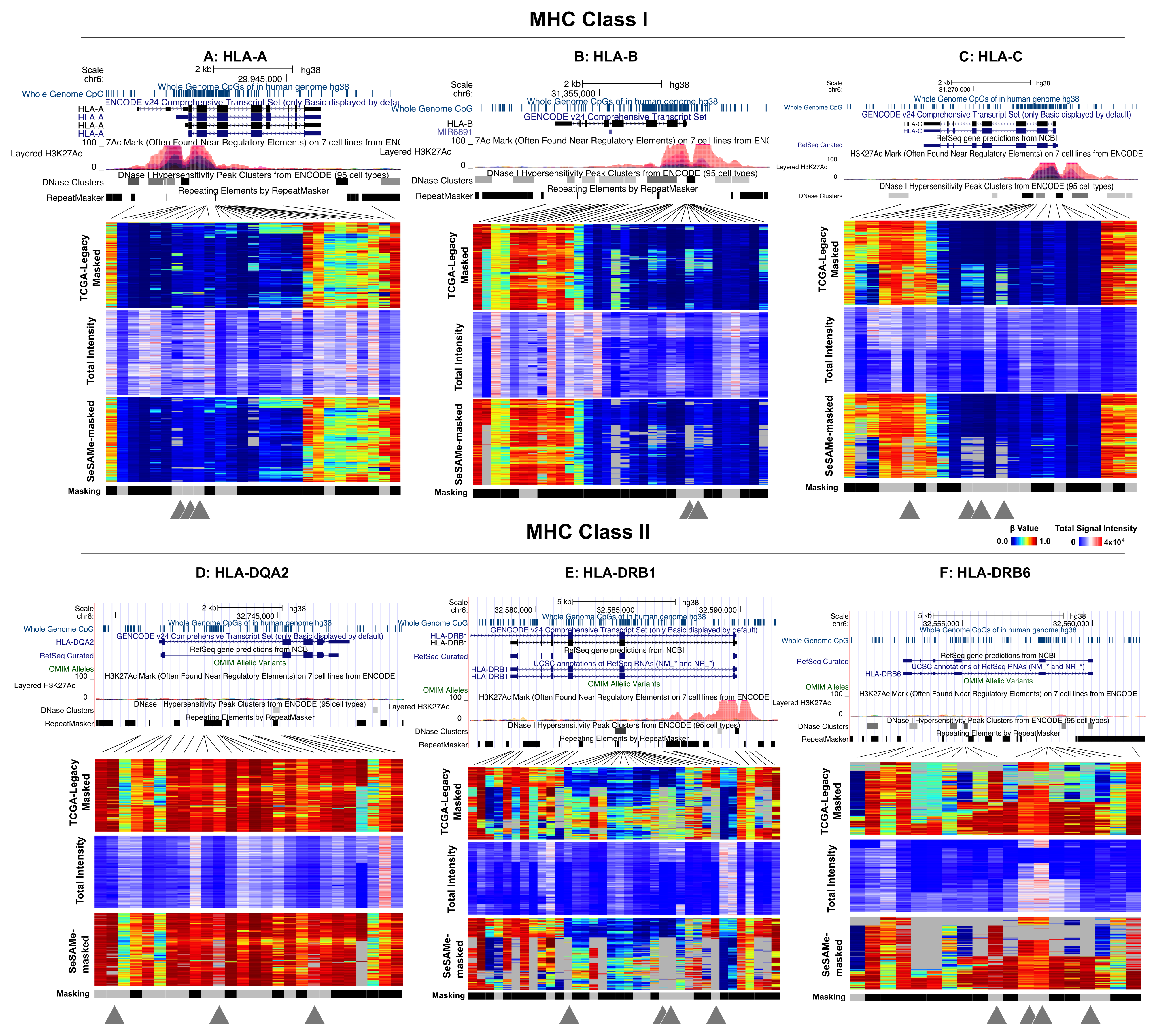
Innovating DNA Methylation Informatics
I llumina Infinium DNA Methylation BeadChips (HM450, EPIC) and high throughput bisulfite sequencing (BS-seq) represent the two most successful and widely used genome-scale DNA methylation platforms of assay. We develop statistical methods and software to control the quality of these experiments and new analytics so that investigators get the most out of their measurements. We design and comprehensively characterize Infinium microarrays and provide the most accurate normalization subroutines and best practice suggestions for analyzing Infinium microarray data. We also explore innovative, unconventional use of BS-seq for genotyping, quality control, and study of somatic genetic abnormalities.
Learn More: Nucleic Acids Res 2017 Nucleic Acids Res 2018