CytoMethIC
C ytoMethIC is a data inference tool and model collection for DNA methylation-based inference.
Learn More: User Manual
SeSAMe
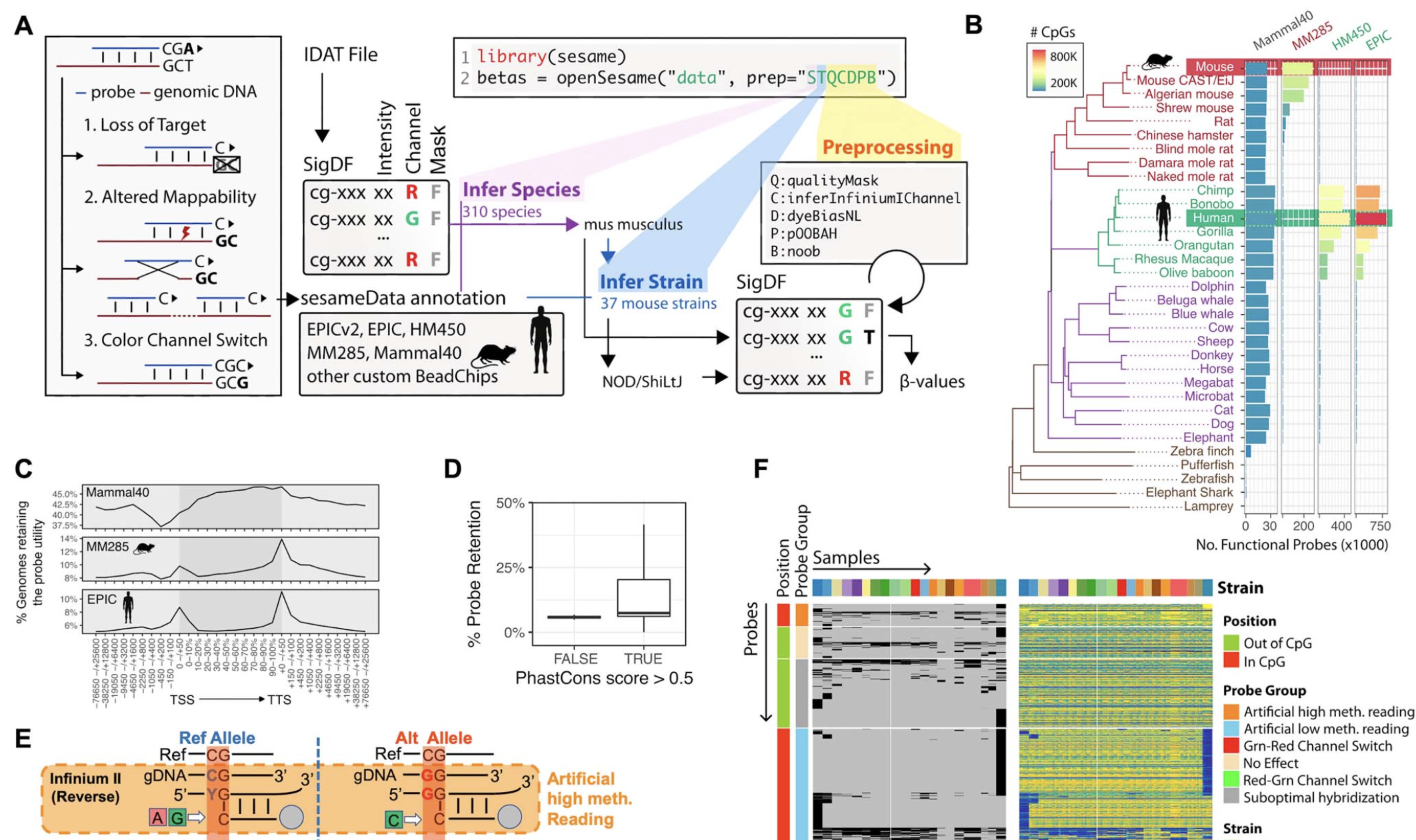
S eSAMe is a R/Bioconductor package for processing Infinium DNA Methylation Microarray. SeSAMe features more accurate detection calling, intelligenet inference of ethnicity, sex and advanced quality control routines.
Documentation:
(developmental)
(1.18)
(1.16)
(1.14)
Bioconductor
Github
Learn More:
Nucleic Acids Res 2024
Brief Bioinf 2023
Cell Genomics 2022
Cell Systems 2019
Nucleic Acids Res 2018
Nucleic Acids Res 2017
BISCUIT
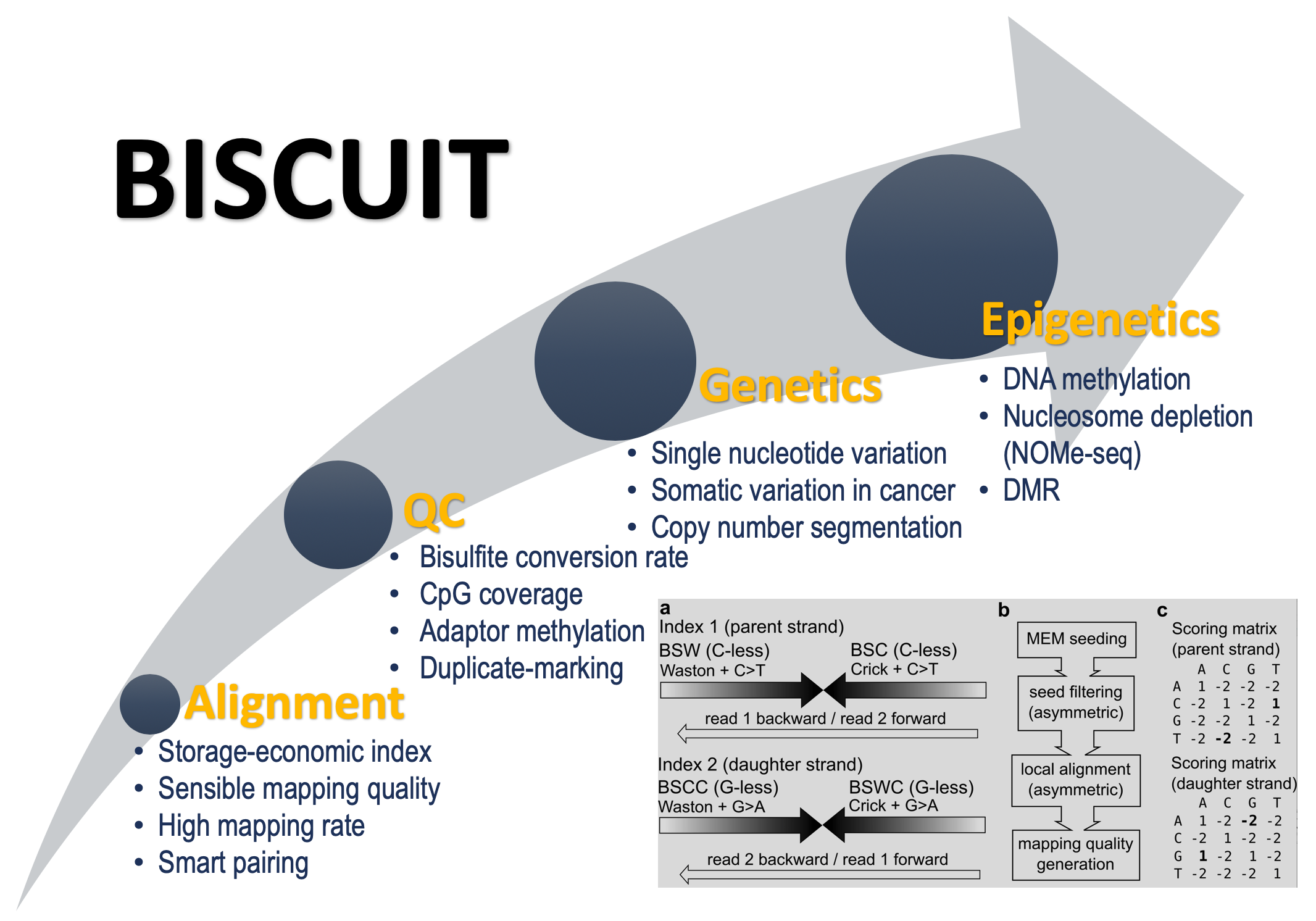
B ISCUIT is a commandline tool for preprocessing bisulfit sequencing data. It's functionalities include short read alignment, read pileup, methylation extraction and downstream analysis. BISCUIT sequence alignment is efficient and accurate. BISCUIT could also call SNPs from BS-seq data. It also supports NOMe-seq.
Learn More: Nucleic Acids Res 2024 User Manual
pyComplexHeatmap
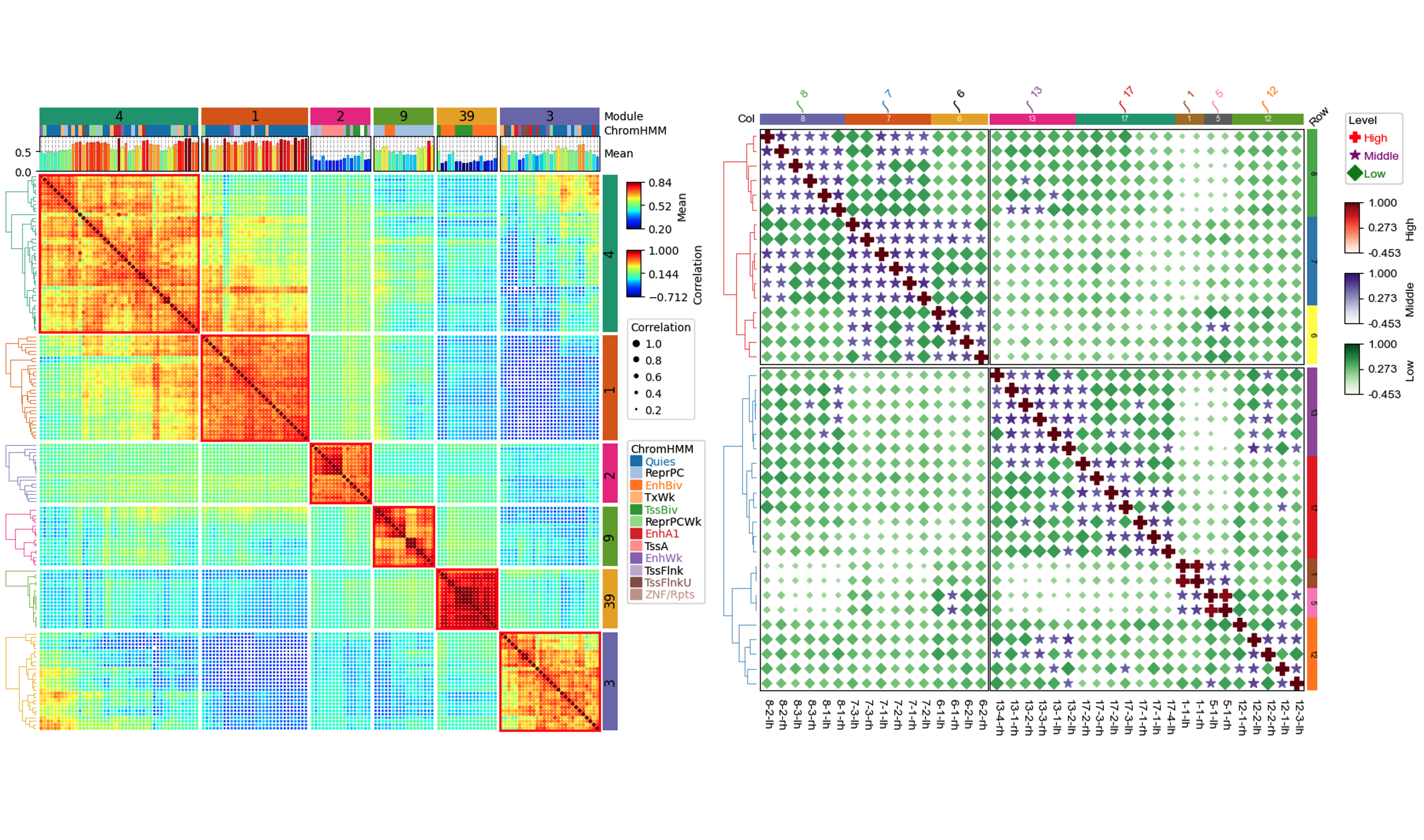
led by Wubin DingP yComplexHeatmap is a versatile and user-friendly Python package, to fill the multidimensional data visualization gap in the Python-based data science ecosystem. We showcased the main features of PyComplexHeatmap in rendering complex biological datasets with rich annotations. PyComplexHeatmap facilitates advanced genomics data analysis in Python, including rendering the OncoPrint plot for cancer genomics data analysis and dissecting single-cell multiomics data of multiple dimensions.
Learn More:
iMeta 2023
User Manual
Github
TransVar
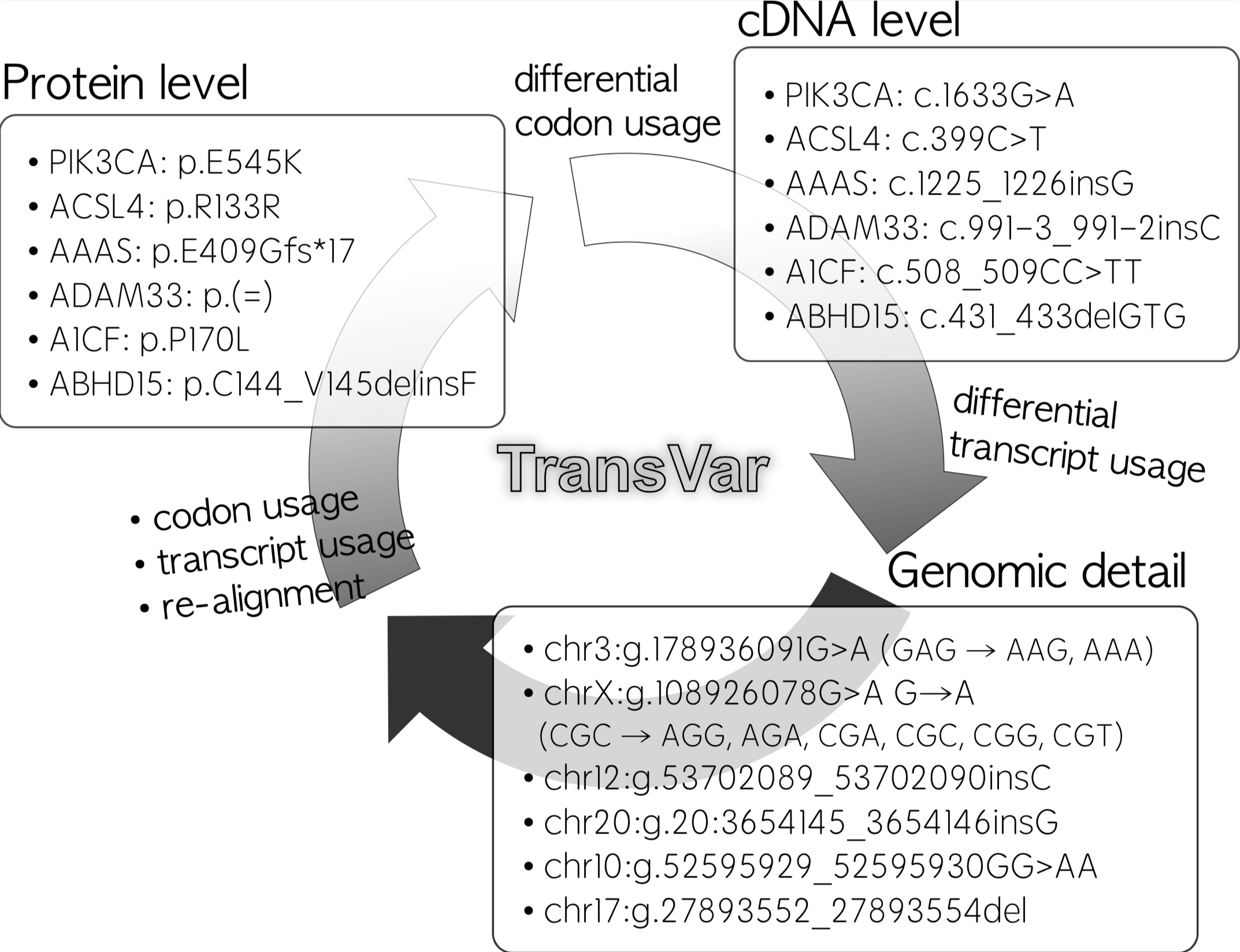
T ransVar is multi-way annotator for genetic elements and genetic variations. It operates on genomic coordinates (e.g., chr3:g.178936091G>A) and transcript-dependent cDNA as well as protein coordinates (e.g., PIK3CA:p.E545K or PIK3CA:c.1633G>A, or NM_006218.2:p.E545K, or NP_006266.2:p.G240Afs*50), and was designed to resolve ambiguous mutation annotations arising from differential transcript usage.
Learn More: Nature Methods 2015 Web Server Github Documentation